Description
Required Software
MATLAB R2013b or above
MATLAB Parallelizing license (optional)
Overview
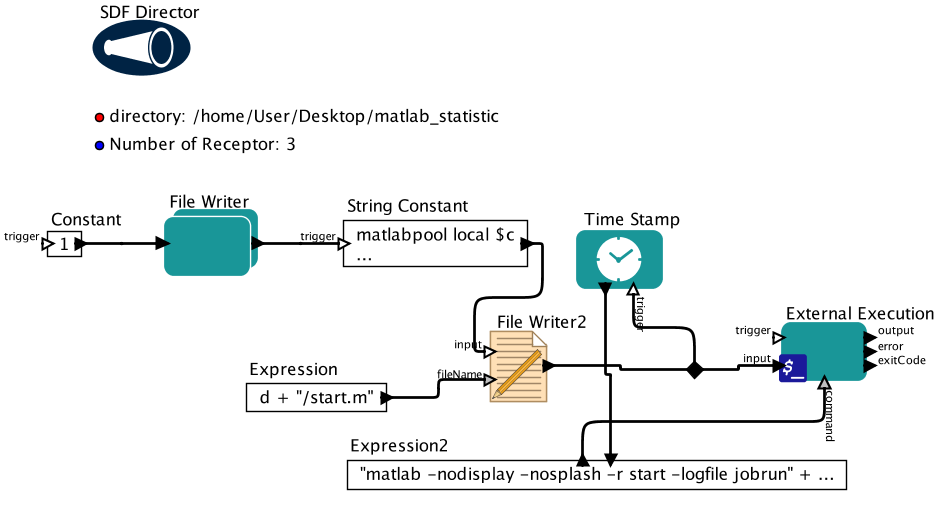
Usage
Parameters
d This is the directory for the input CSV files and the output files.
n Number of ensemble receptor structures. For example, if user wants the best 2ensemble receptor structures out of the 8 structures they used for virtualscreening, they will specify 2.
high Name for the CSV file that contains 999999 to represent non-binding events.
zero Name for the CSV file that contains 0 to represent non-binding events.